Tim Sterne-Weiler, PhD
Senior Principal Scientist, Computational Biology & Translation
Genentech
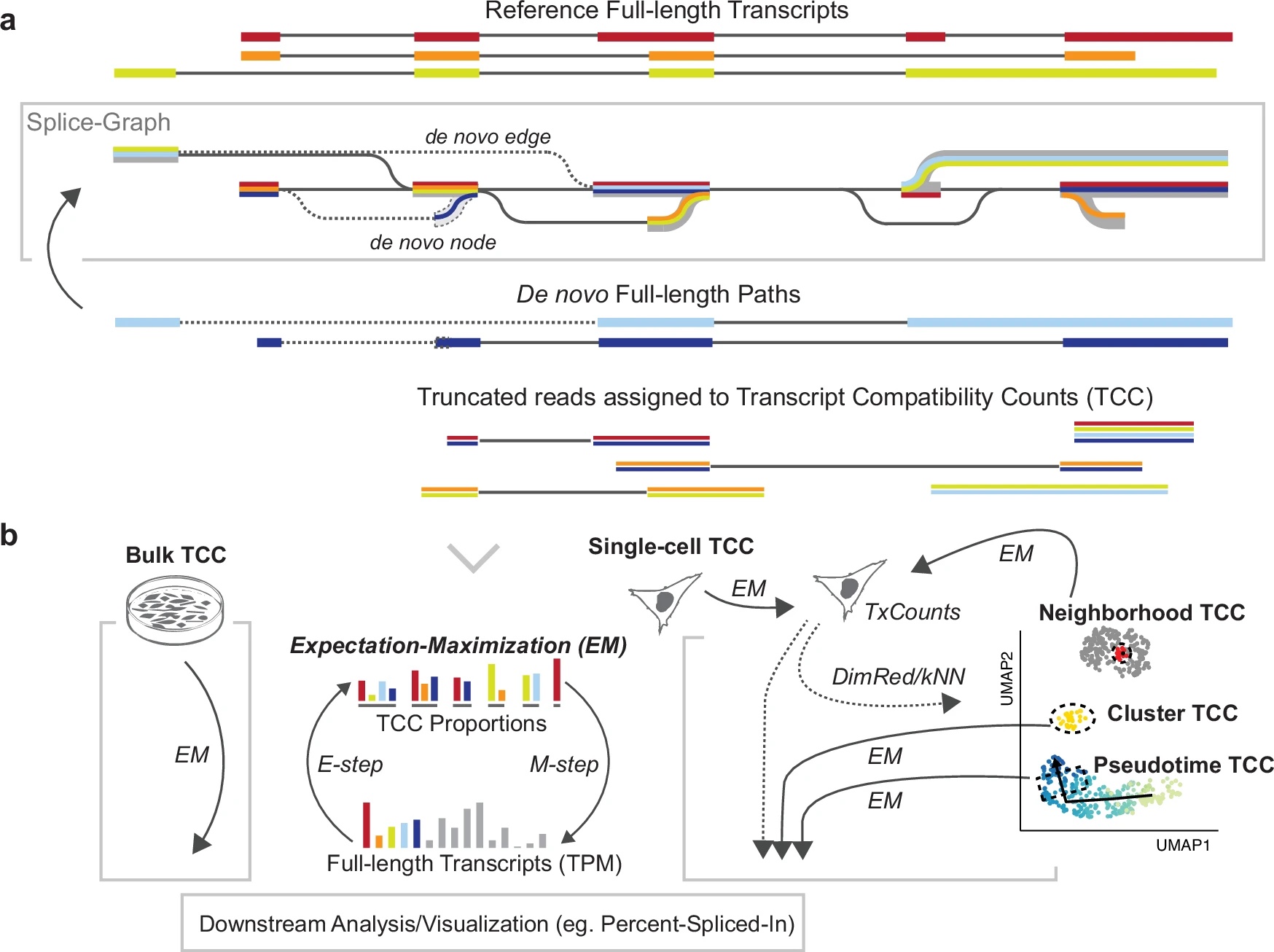
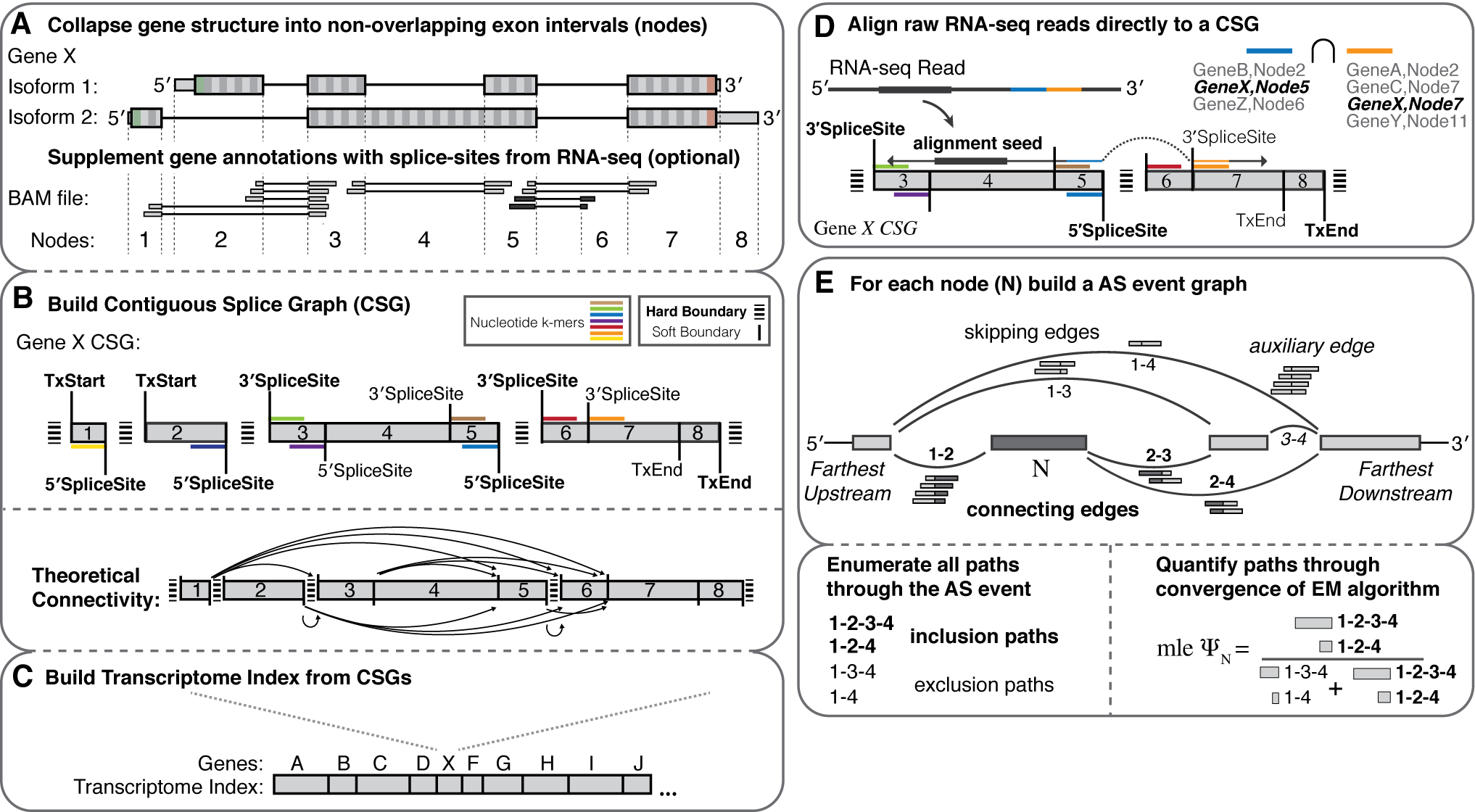
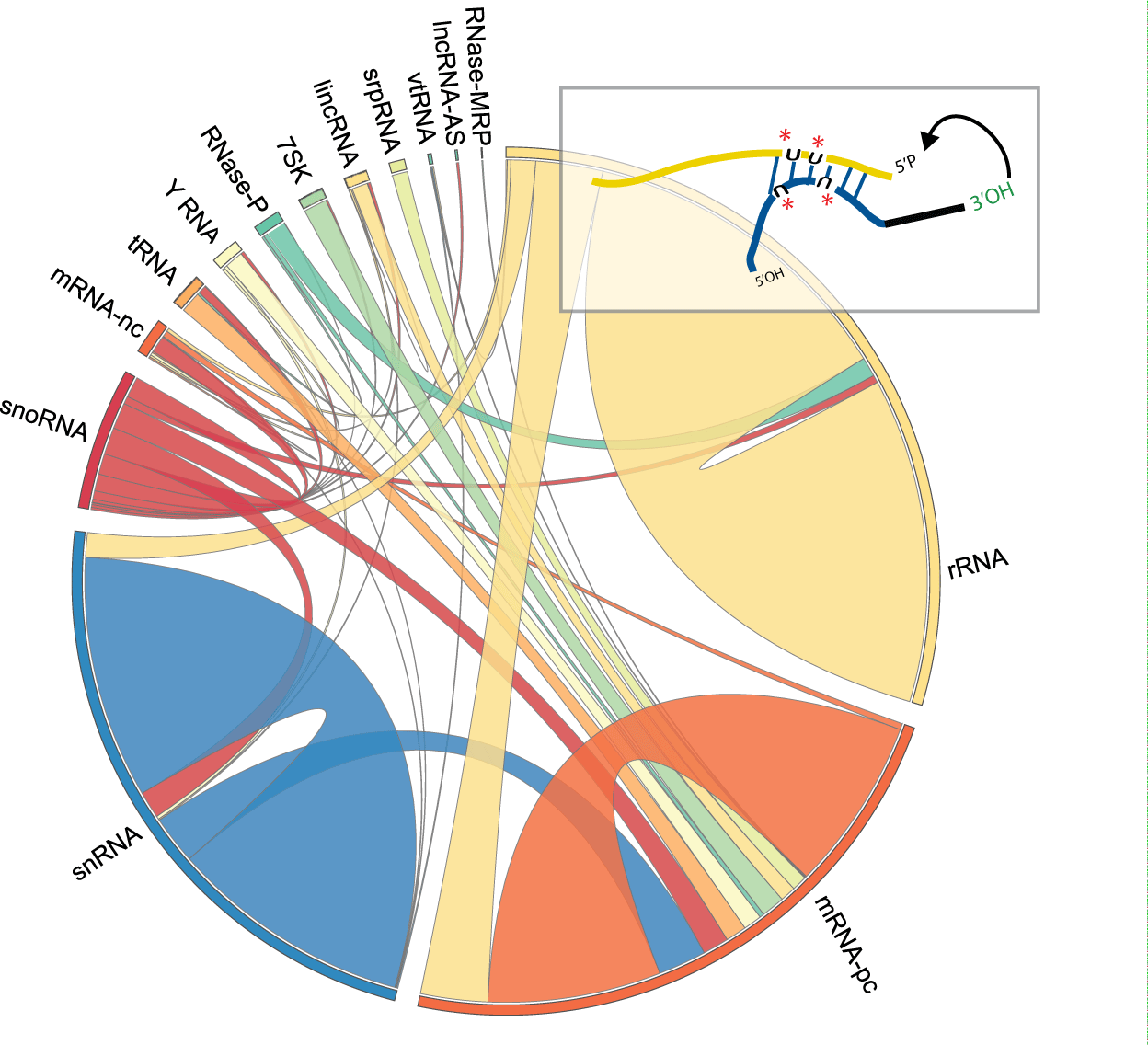
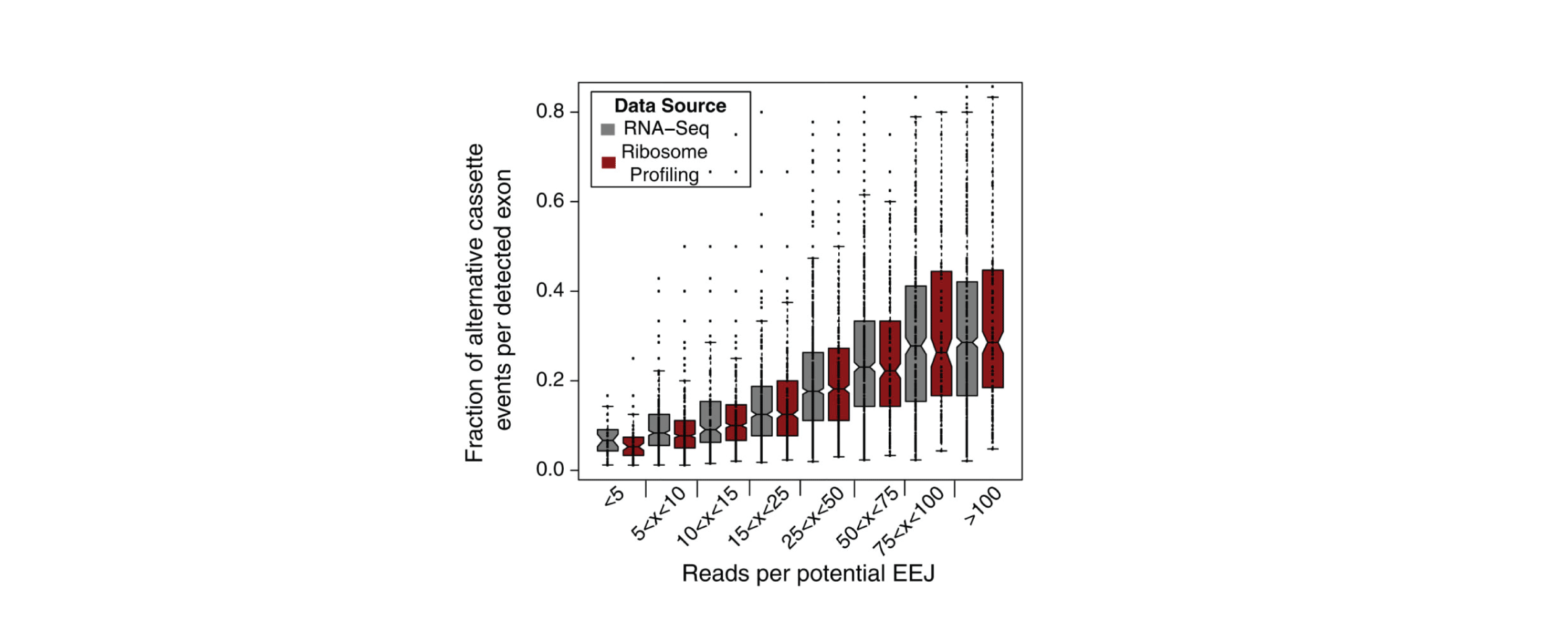
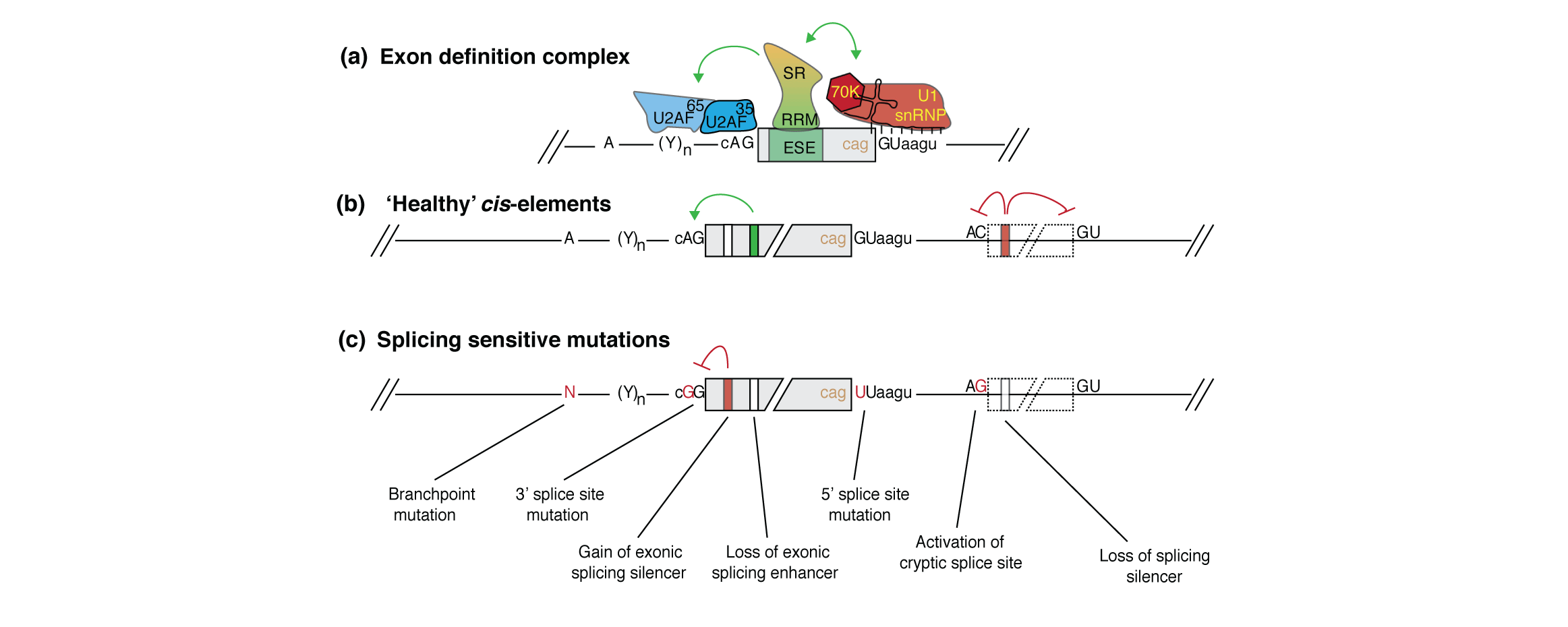
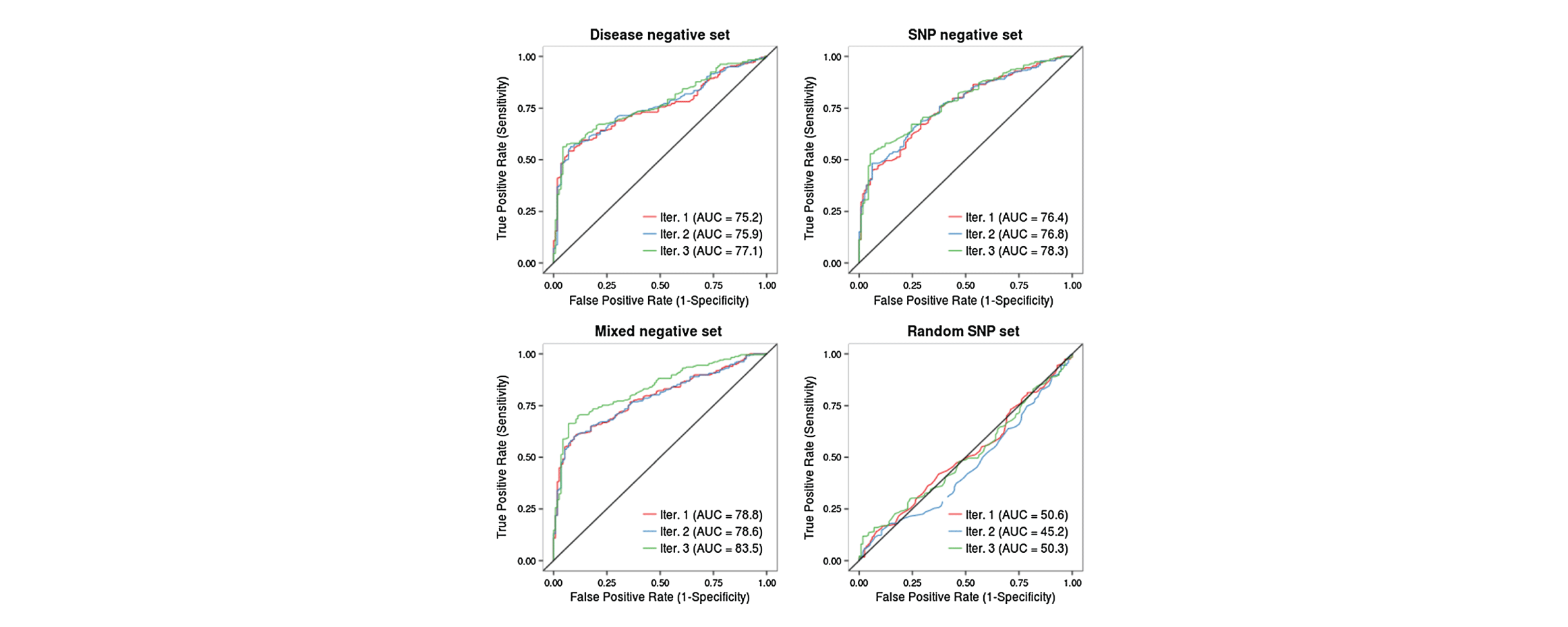
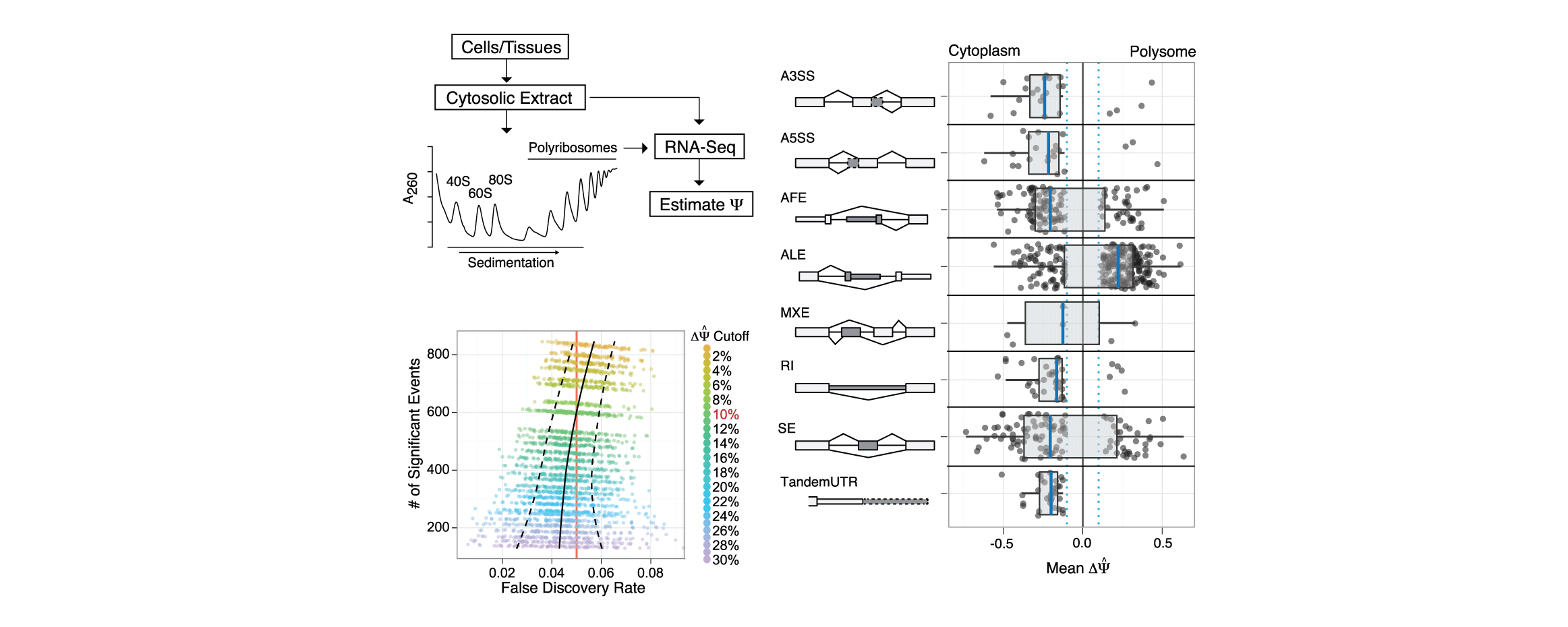
My research interests lie at the intersection of computational genomics, machine learning and human disease. I'm currently a Principal Scientist at Genentech, studying gene dysregulation in cancer. Formerly a postdoc in the Blencowe lab at the University of Toronto.